Machine learning reduces microscope data processing time from months to just seconds

Ever since the world's first ever microscope was invented in 1590 by Hans and Zacharias Janssen --a Dutch father and son-- our curiosity for what goes on at the tiniest scales has led to development of increasingly powerful devices.
Fast forward to 2021, we not only have optical microscopy methods that allow us to see tiny particles in higher resolution than ever before, we also have non-optical techniques, such as scanning force microscopes, with which researchers can construct detailed maps of a range of physical and chemical properties.
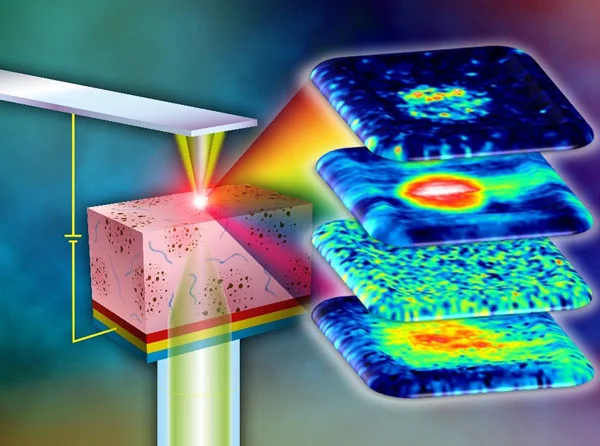
IBEC's Nanoscale bioelectrical characterization group, led by UB Professor Gabriel Gomila, in collaboration with members of the IBEC's Nanoscopy for nanomedicine group, have been analysing cells using a special type of microscopy called Scanning Dielectric Force Volume Microscopy, an advanced technique developed in recent years with which they can create maps of an electrical physical property called the dielectric constant.
Each of the biomolecules that make up cells --that is, lipids, proteins and nucleic acids-- has a different dielectric constant, so a map of this property is basically a map of cell composition. The technique that they developed has an advantage over the current gold standard optical method, which involves applying a fluorescent dye that can disrupt the cell being studied.
Their approach doesn't require the addition of any potentially disruptive external agent. However, the application of this technique requires a complex post-processing process to convert the measured observables into the physical magnitudes, which for eukaryotic cells involves huge amounts of computation time. In fact, it can take months to process just one image in a workstation computer, since the dielectric constant is analysed pixel by pixel using local reconstructed geometrical models.
Months to seconds
In this new study, published in the journal Small Methods ("Fast Label-Free Nanoscale Composition Mapping of Eukaryotic Cells Via Scanning Dielectric Force Volume Microscopy and Machine Learning"), the researchers opted for a new technique to speed up the microscope data processing. This time, they used machine learning algorithms instead of conventional computational methods.
The result was drastic: once trained, the machine learning algorithm was able to produce a dielectric biochemical composition map of the cells in just seconds. In the study, no external substances were added to the sample, a long-sought goal in the composition imaging in cell biology. They achieved these rapid results by using a powerful type of algorithm called neural networks, which mimics the way that neurons in the human brain operate.
The study was first-authored by Martí Checa, who carried it out as part of his PhD in Gomila's group at IBEC. He is now a postdoctoral researcher at the Catalan Institute of Nanoscience and Nanotechnology (ICN2). "It is one of the first studies to provide such a rapid label-free biochemical composition map of dry eukaryotic cells", Checa explains.
Indeed, in this proof-of-concept study, the researchers used dried out cells, to prevent the huge effects of water in the dielectric measurements due to its high dielectric constant. In a recently published follow-up study (Nanomaterials, "Dielectric Imaging of Fixed HeLa Cells by In-Liquid Scanning Dielectric Force Volume Microscopy "), they also analysed fixed cells in their natural in-liquid state. Here they were able to compare the values obtained in the dry and liquid models in order to render an accurate map of the biomolecules that make up eukaryotic cells. These are the multi-structured cells that animals, plants, fungi and other organisms are composed of.
"The next step in this research is to apply the method to electrically excitable living cells, such as neurons, where intense electrical activity occurs. We are excited to see what can be obtained with our technique in these systems" Prof. Gomila adds.
Biomedical applications
The researchers validated their methodology by comparing their findings to well-known facts about the composition of cells, such as the lipid-rich nature of the cell membrane or the high quantity of nucleic acids present in the nucleus. With this work, they have opened up the possibility of analysing large quantities of cells in record time.
This study is expected to provide an invaluable tool to biologists to conducting basic research, as well as, to open up potential medical applications. For example, changes in the dielectric properties of cells are currently being studied as possible biomarkers of some illnesses, such as cancer or neurodegenerative diseases.
"It is the first study to provide such a rapid nanoscale biochemical composition map from dielectric measurements of dry eukaryotic cells, which are classically seen as being extremely difficult to map due to their complex three-dimensional topography", declares Martí Checa, first author of the paper.